Animals are made up of many different types of cells, like neurons or muscle cells, which have very different functions. These different cell types are generated during embryonic development, differentiating and continuing to have different function because they make different proteins. However, an animal’s cells all have the same DNA, which contains the instructions for makings those proteins, so each cell type must use a different subset of these instructions to make only the proteins that they need. The cell turns on the DNA it needs by first transcribing it into messenger RNA, or mRNA, and then translating that mRNA into protein. To study how different cell types emerge during development through the action of different DNA sequences, we visualize mRNAs in fruit fly embryos.
We study fruit flies because they have a surprising amount in common with humans and are easier and more ethical to work with. Many of the proteins found in a fruit fly are also present in humans, and many of the ways that cell types are generated are similar between these two animals. Therefore, studying mRNA production and development in fruit flies can help us learn about how the process works in humans and why it sometimes goes wrong. Scientists have been studying fruit flies for decades, so there are many experimental tools available for us to visualize mRNA and to genetically modify fruit flies.
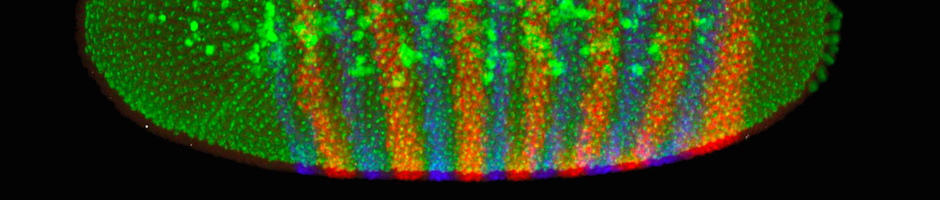
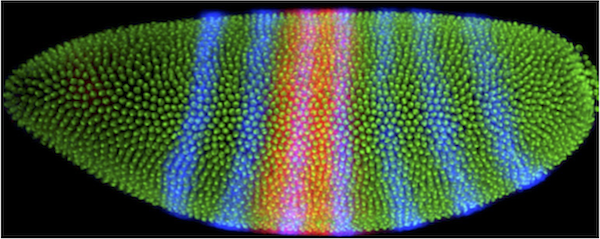
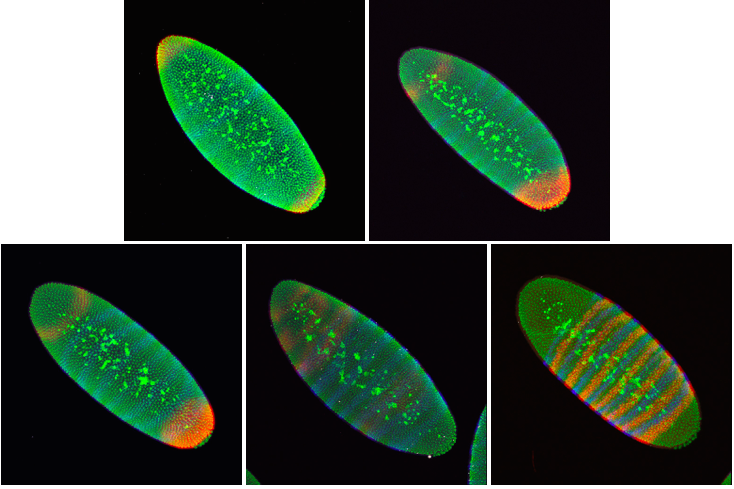
A fruit fly develops very quickly. Just three hours after the egg is fertilized, the original single cell has divided 14 times, yielding about 6000 cells. The embryos at this stage look like tiny footballs, with all of their cells on the surface and yolk in the middle. The cells look similar, but they actually already know what part of the body they will become. We know this because embryos lacking specific proteins at this stage develop into larvae with missing body parts. For example, embryos missing the protein produced by the mRNA shown in red below are missing the middle part of their body. In contrast, embryos missing the protein produced by the blue mRNA are missing every other segment of their body. Therefore, knowing where and when mRNAs are produced in an embryo can tell us about their specific functions.
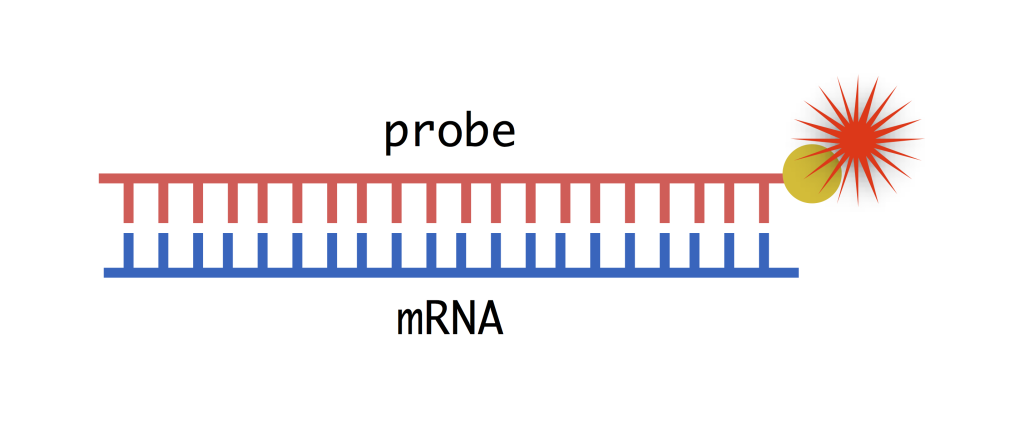
So how do we visualize where the mRNAs are in the embryo? Unlike DNA, mRNA is single stranded, but that single strand can bind very tightly to another single strand with a complementary sequence (A binds with T, C binds with G). We can artificially synthesize this complementary mRNA strand, which we call a probe, and label it for visualization (more on that below). To make a probe, all we need to know is the sequence of the mRNA, which we can get from the original DNA sequence of the gene whose mRNA we want to visualize. The process of seeing where mRNA is expressed in an organism using probes is called in situ hybridization. Hybridization refers to the binding of the mRNA and the complementary probe, and in situ means that we do the experiment “in place”– meaning in the embryo. Below is the protocol for using in situ hybridization to visualize mRNAs in fruit fly embryos.
- First, we place approximately 2000 male and female fruit flies in a cage that allows us to collect their embryos. We put an agar-molasses plate on the bottom of the cage, where the flies will deposit their eggs.
- We change the plates after four hours to ensure that most of the embryos are at the developmental stage that we study. The plates are covered in embryos (each white speck to the right) that we flush with water into a small bucket with a mesh bottom for collection.
- Once the embryos are collected in the bucket, we can put them in bleach to release the eggshell (yes, fruit flies have eggshells!). We then shake the embryos in a formaldehyde-heptane mixture for 25 minutes to stop the cells from dividing and permanently freeze the embryos at the right age. Vigorously shaking the embryos in a methanol wash will then pop off their outer membrane, resulting in naked embryos. Subsequent methanol and ethanol washes ensure that the embryos are in a pure ethanol environment in which they can be frozen until they’re needed for an experiment.
- Our in situ hybridization protocol involves a week of moving tiny amounts of liquid in and out of the tube holding our embryos. This process begins with adding the complementary probes that bind the mRNAs we’re interested in and then washing away the unbound excess probes.
- The probes aren’t visible on their own, so we need to attach a molecule that produces light to them. This is a multi-step process. Initially we synthesize the mRNA probe with an extra chemical attachment at the end. That chemical attachment can bind to a protein that we add after the probe is bound to the mRNAs. That protein can then catalyze a reaction that deposits a fluorescent dye. We can deposit two different fluorescent dyes (red and blue) to visualize two mRNAs at once. After we wash with the protein and then the dye, the dye is stuck in place (in situ!) We also stain the embryos with Sytox, a chemical that stains all DNA and fluoresces green, allowing us to see not just the mRNAs, but also each of the 6000 cells that form the embryo.
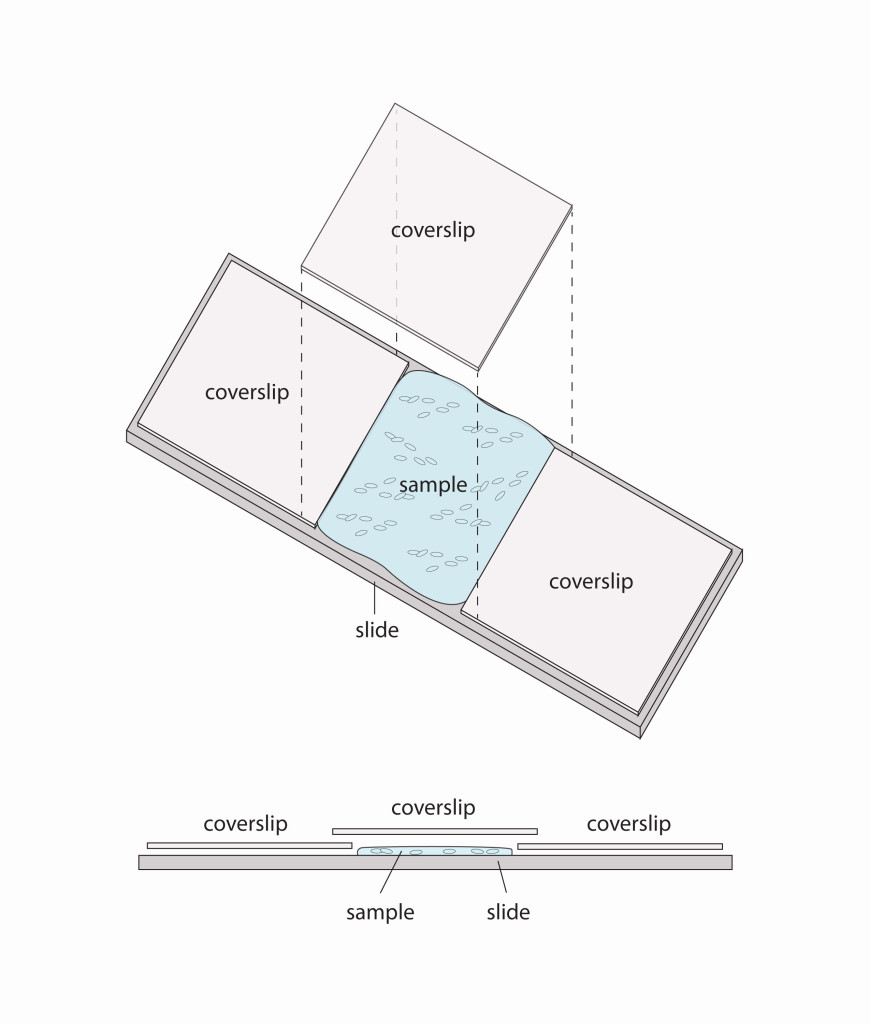
- After in situ hybridization, we are ready to prepare the embryos for mounting on slides. The first step is to clear the embryos with an organic chemical that makes the yolk transparent and allows us to image the embryo from top to bottom. Then we simply spread the embryos onto glass slides and cover them in DEPEX, a viscous liquid that suspends the embryos and protects them from getting squished by the coverslip. We also lay two more coverslips on either side of the patch of embryos to act as legs of a bridge to the overlaying coverslip, further protecting the embryos.
- The DEPEX dries for 5 days and then the slides are ready to be imaged. Our microscope is a laser scanning microscope, which scans a laser across the embryo to generate a high-resolution image pixel-by-pixel. We need high resolution to distinguish the tiny cells that make up the embryo. The microscope is controlled by a computer with software that records where the embryos are on the slide. We also record the top-most and bottom-most edge of each embryo. After we have found all the embryos on a slide, the software tells the microscope to go back to each embryo and take a “stack” of 2D images, from top to bottom. These images, when combined, create a 3D image of each embryo.
- We take 3D images of many embryos. For a single mRNA of interest, we take a lot of pictures so we can average the mRNA pattern and reduce the experimental uncertainty of our measurements. Also, there are many different mRNAs that are important for the developing embryo, so we do many in situ hybridizations to learn about all of them.



The DePace lab is in the Systems Biology department at Harvard Medical School. The lab studies the mechanism and evolution of transcription in animals, using fruit fly embryos as a model system. This protocol was written by Benjamin Vincent, Francheska Lopez-Rivera, Jeehae Park, Max Staller, Meghan Bragdon, and Zeba Wunderlich.