A virus races through densely populated China, and the numbers are grim: mortality rates are north of 80%, and millions are dead in the first year of the outbreak. Three years later, the epidemic reaches the United States, and spreads to 16 states in three months. DNA sequencing proves that the American version of the virus originated in China. Once in the U.S., the virus is rapidly disseminated by contaminated transportation infrastructure. With no known cure for the virus, the focus shifts to isolating known cases while the epidemic runs its course.
This devastating epidemic is very real and ongoing. First identified in the United Kingdom in the 1970s, today’s Porcine Epidemic Diarrhea Virus (PEDV) outbreak began in China in 2010. We are still learning the extent to which our globalized infrastructure helped this epidemic spread. In the meantime, PEDV has driven US pork prices up more than 13 percent in the first year of the epidemic. To offset the loss of pigs killed by PEDV, growers are increasingly relying on imports from Europe. While this solution is only temporary, it has helped lessen the impact of PEDV on retail pork prices, raising little concern from consumers.
Fortunately, PEDV does not appear to infect humans, but it is a close cousin to other viruses that have made the jump across species boundaries including Severe Acute Respiratory Syndrome (SARS) and Middle East Respiratory Syndrome (MERS). Each infected organism (animal or human) offers a virus another billion chances to evolve and change. The H5N1 and H7N9 “bird” flus as well as H1N1 “swine” flu first infected workers caring for birds and pigs. These flu strains have an unusually high mortality rate, but appear to not spread effectively between people; in these cases, the animals act as “reservoirs” for the disease. In public health circles, this realization has led to the “One Health” concept, which acknowledges that managing and treating disease in animals is as important as preventing disease in humans.
Early understanding of the link between animal and human health revolutionized public health hundreds of years ago, when livestock were moved outside of people’s homes. Today, however, modern infrastructure has counter-intuitively strengthened the link between animal health and human health: more people means more domestic livestock, and global trade networks offer ample opportunities for the spread of disease. Furthermore, today’s Ebola epidemic has vividly illustrated that the spread of disease through urban centers has global consequences. Ebola outbreaks, which are thought to originate from bats, may be more difficult to limit now due to increasing urbanization. Both globalization and climate change may be contributing to the spread of insect-borne diseases such as West Nile virus and Chikungunya virus, which were once thought to be restricted to tropical regions.
Animal populations also play a major role what many consider to be one of the greatest health threats in the 21st century—the growing ineffectiveness of antibiotics, one of the 20th century’s greatest health advancements. Clinical overuse of antibiotics has played a well-known role in the rise of antibiotic resistance. Somewhat less well-known is that most antibiotics manufactured every year end up in animals instead of humans. The FDA estimates that about 8.3 million kilograms of “medically important” antibiotics (meaning drugs similar to or identical to antibiotics used for humans) were used in food-producing animals. Over the past half-century, antibiotics have been added to the feed of healthy farm animals. For reasons that are not entirely clear, this helps animals grow more quickly. Efforts to curb this practice, particularly in the European Union, have faced many challenges: antibiotics are cheap, and the conditions in modern farms often lead to animals getting sicker when antibiotics are removed. In the meantime, the effectiveness of antibiotics in the clinic is undergoing a steady decline.
Keeping up with these emerging health threats may require radically new approaches to drug and vaccine development. Complete physical containment of biological threats that have reached animal populations is impossible; improving farm practices and human behavior doesn’t prevent the spread and evolution of diseases in wild animals. As an alternative to physical containment, researchers are exploring the potential for biological containment: can we engineer biological systems that limit the spread of disease in animal reservoirs? If so, we could fight diseases when they occur in animals, improving the health of livestock and limiting animal-to-human disease transmission.
The field of synthetic biology [full disclosure: my area of research] may offer new approaches to managing diseases that spread through animal populations. Synthetic biologists are biological engineers: the world around us provides countless examples of how biology is a unique and powerful engineering substrate. It’s hard to imagine a micron scale robot that derives all of its power from the local environment and can make copies of itself every 30 minutes, and yet trillions of bacteria in your gut do just that. Synthetic biologists hope to tap into the power of biological systems to make new medicines, vaccines, chemicals, foods, and all the other products that biology is good at making.
Simply developing new tools for engineering biology is not enough to enable new approaches to fighting disease. These tools literally need a place to live: synthetic biologists are engineering organisms that can thrive in specific niches, including the gut microbiome. In people and animals, one of the richest niches for microbes is the gut. The trillions of microbes living in your gut aid in digestion and help protect us from the few microbes that can make us sick. How might engineered bacteria in the gut help limit the spread of infections?
Gastrointestial infections like PEDV are often transmitted through the “fecal-oral” route: food or water contaminated by the feces of a sick individual. Bacterial hemorrhagic diarrhea, like that caused by E. coli O157:H7, is often acquired through meat tainted in farms and slaughterhouses. Engineered microbes could help protect us from these infections before they make us sick. Better yet, livestock “vaccinated” with an engineered microbiome of their own could block these bad microbes at the source: farm animal feces.
Engineered microbiome research is still in its early stages. The first step towards microbiome treatments, fecal transplants, have had significant success in early clinical trials—good microbes from a healthy donor can “boot up” a functioning gut microbiome to crowd out the bad microbes that were causing infection. The biggest role for fecal transplants today is to help patients recover from Clostridium dificile infections. C. dificile can take over the gut when other competing microbes have been wiped out by antibiotics; fecal transplants can supply enough good microbes to take that territory back. Of course, it would be better to avoid gut infections in the first place. Synthetic biology researchers have also been able to engineer normal gut bacteria to sense changes in the gut environment in mice. Advanced versions of these engineered bacteria could someday stand guard in your gut and administer therapeutics if an infection or other problem is detected.
Synthetic biologists are also exploring how engineered microbes may be able to protect against antibiotic resistance. Bacterial infections are not always caused by “bad” bacteria; many infections (including E. coli O157:H7) are caused by “good” gut bacteria that have acquired bad genes. Genes conferring antibiotic resistance in particular can spread rapidly through populations, passing between different individuals in a microbial population on mobile genetic elements. Early experiments with engineered proteins that can target and cut specific DNA sequences have shown that it may be possible to target and eliminate these antibiotic resistance genes from a population—stopping the spread in its tracks.
Microbiome treatments may transform how we treat disease in people and farm animals, but there will always be diseases that incubate and evolve in wild animals. In the case of insect-borne diseases like malaria, newly published research has shown that a naturally occurring species of bacteria living in the gut of mosquitoes might be able to prevent the malaria parasite from infecting the mosquitoes. Feeding mosquitoes probiotic nectar with this species significantly reduces their susceptibility to the infection, making them less likely to spread malaria to humans.
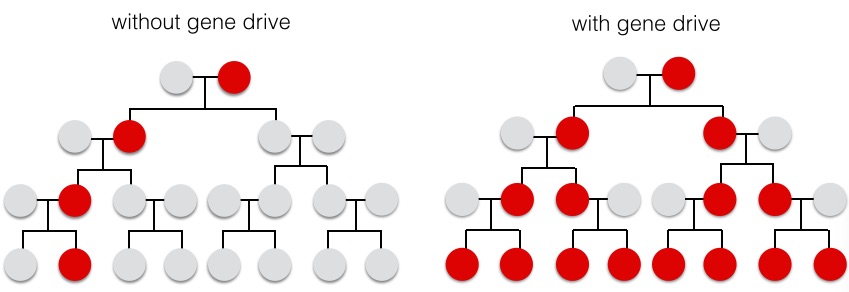
Another radical (yet increasingly feasible) approach to managing insect-borne disease is to engineer the mosquito populations themselves through genetic elements known as “gene drives.” Like viruses, gene drives are selfish genetic elements. While normally offspring receive one copy of each gene from the father, and a second copy of each gene from the mother, with gene drives, the gene in question can duplicate itself and erase the copy of the gene from the other parent—allowing a new gene to spread through a population much more rapidly.
In the case of malaria, mosquitos engineered with gene drives that cause sterility in offspring could mate with wild mosquitoes. As the sterility genes spread, the mosquito population would decline, and eventually be eliminated from the ecosystem. Others have proposed designing gene drives that protect the mosquito from being infected with the Plasmodium parasite that actually causes malaria, leaving mosquito populations intact.
Gene drives have not been tested in the wild yet, but tools for controlling mosquito populations have undergone limited field trials. One technique involves the release of sterile male mosquitos into wild populations; these males mate with wild females and reduce the number of offspring in the next generation. This has been done for decades via the use of radiation or chemicals to sterilize the males; over the past decade more precise bioengineering tools have lead to engineered male mosquitos with more predictable traits.
The development of this technology is illustrative of the rapid pace at which biotechnology is advancing. The gene editing technologies essential to building gene drives are the same as those used to target antibiotic resistance genes in the microbiome. This technology was first described in early 2013; about a year later, papers extending this technology to microbes, plants, and animals have been published, including one where researchers in China had successfully edited the genomes of monkeys. The cost of reading and writing DNA has become so cheap that new bioengineering advances can spread virally (in the internet sense of the word), with researchers synthesizing new genetic circuits based on the latest advancements around the world. This ease of information sharing and DNA synthesis represents a fundamental change in how we do science, and how quickly new techniques and designs can disseminate among researchers.
Caught between the evolving threat of animal-borne disease and the rapid development of biotechnology to meet these threats lies a patchwork of regulations designed for 20th century biology. In the United States, many regulations are based on the control of agricultural pests or diseases and were not originally intended to regulate engineered organisms. For example, if you want to engineer plants, the tools you use (rather than the changes you make to the plant genome) determine who regulates your new plant. Many researchers use a bacteria called Agrobacterium, which is good at taking a specific piece of DNA that you give it and transferring it into the plant of your choice. If you want to sell the engineered plant, the USDA’s Animal and Plant Health Inspection Service (APHIS) has regulatory jurisdiction because wild Agrobacterium is a plant pathogen. If you use a gene gun (literally a gun that shoots DNA into things) to put the same piece of DNA into the same plant, you are exempt from APHIS oversight. This theme is repeated across the regulatory system, with policies generally concerning specific methods rather than the end result. In today’s world, where new methods develop in a span of months rather than decades, these regulations quickly become outdated.
Gene drives and other environmental applications of synthetic biology are likely to cross regulatory and international borders. A gene drive trial in the United States could conceivably fall under the jurisdiction of the FDA, USDA, and EPA. Internationally, field trials with engineered mosquitos have been performed under geographic isolation (usually on islands) and under the specific laws of the local country. A gene drive-based solution could have a larger geographical range, requiring multinational cooperation for regulation and oversight.
Of course, regulations do not prevent people from engineering biology. As with other technologies, advances in biotechnology are becoming increasingly accessible to the general public in addition to skilled researchers. While you can’t exactly build a gene drive in your basement, there is a growing community of hobbyists in the synthetic biology world. To adapt to this leveling playing field, we will need a better understanding of the risks posed by environmental deployment of biotechnology, and cultivate communities that encourage responsible technology development. Fortunately, these issues are being discussed, with efforts to include input from all of the relevant stakeholders. For example, the Synthetic Biology Project, led by the Woodrow Wilson International Center for Scholars, has convened a series of workshops that bring synthetic biologists, environmental scientists, regulators, hobbyists, and concerned NGOs together to discuss new case studies. These case studies are important in focusing the discussion to specific projects that are likely to be field-tested in the near to medium term; separating the real elements of new technology from the hype (both positive and negative) is essential to making informed decisions. These case studies also help the synthetic biologists involved in these projects to take input from stakeholders, and adjust the design of their project to better address their concerns. In a series of publicly available reports, the Synthetic Biology Project has outlined research and regulatory priorities relevant to helping us understand and evaluate new synthetic biology technologies.
Improving human health in the 21st century will not be driven entirely by biotechnology—culture, politics, and values play a large role in how we manage disease—but we won’t be able to simply rely on the drugs and practices that got us here. Given the myriad emerging health threats we face, it will be a challenge to maintain the current health standards of the developed world over the course of this century. Antibiotic resistance can’t be addressed by continuing our current widespread use of antibiotics in agriculture; we will likely need a mix of new practices and technology to save the antibiotics we have left. PEDV has shown us that borders and high local standards don’t shield us from global epidemics. Improving human health in the future will mean improving global health.
While we face immense challenges, we also have unprecedented access to tools and data that can help us adapt to these new realities. Gene sequencing and data sharing enabled pig growers and researchers to trace PEDV to its source and slow the spread of the epidemic. Synthetic biology could allow us to vaccinate pig populations against future PEDV outbreaks, either via genome editing or a microbiome upgrade. Careful and regulated tests of these technologies will also give us a better understanding of how diseases spread and persist, since we will need to adequately quantify the effectiveness of novel interventions. Developing tools that can address human diseases in animal populations would be a breakthrough on par with the discovery of antibiotics: a tectonic shift in how we fight infectious disease.